This document shows semantic search on Elasticsearch on PDF documents without any additional client code.
We use the following features of Elasticsearch:
- Ingest pipelines
- PDF text-extraction
- Inference endpoint
- ELSER sparse-vector text-expansion model
Both ingestion pipeline and inference endpoints require Platinum license or above.
Intended Benefits
Elasticsearch now supports running custom ingestion pipelines. It can handle many different transformations by itself, but also supports custom pipeline steps. Elasticsearch supports running ML models for semantic search and hybrid search.
If Elasticsearch can run the full ingestion pipeline for Delta3, that can significantly reduce the complexity of Delta3 code that runs the pipeline.
On the query side, if Elasticsearch can do semantic search and hybrid-search (including re-ranking), that can reduce the complexity of Delta3 as well.
Ingesting a PDF on Elasticsearch
Next, we describe running a canonical pipeline that involves reading a PDF into Elasticsearch and running semantic search on it, with Elastic cloud. This requires no code on our end, and is all part of the Elastic distribution.
Step 1: Defining an ingest pipeliine
The following pipeline has two steps, called “processors” by Elasticsearch:
- It takes a PDF document and extracts text from it using Tesseract
- It then passes the text to built-in ELSER model to extract embeddings
PUT _ingest/pipeline/pdf-ingest-pipeline
{
"processors": [
{
"attachment": {
"field": "data",
"properties": [
"content",
"title"
],
"remove_binary": true
}
},
{
"inference": {
"model_id": ".elser-2-elasticsearch",
"input_output": {
"input_field": "attachment.content",
"output_field": "content_embedding"
}
}
}
]
}
We extract two fields from the PDF coming in through the “data” field, going into “title” and “content”. We also remove the original binary. By default, Elasticsearch extracts all fields and retains the binary. It places the extracted fields under “attachment” top-level field by default. Note that the PDF is base64-encoded in “data” field; Elastic provides options to pass the binary directly if we want to be efficient. I have not tried it yet.
Elastic provides many “processors” for a pipeline. Here is a partial view. It also allows for custom processors, which I have not yet explored. Reference documentation.
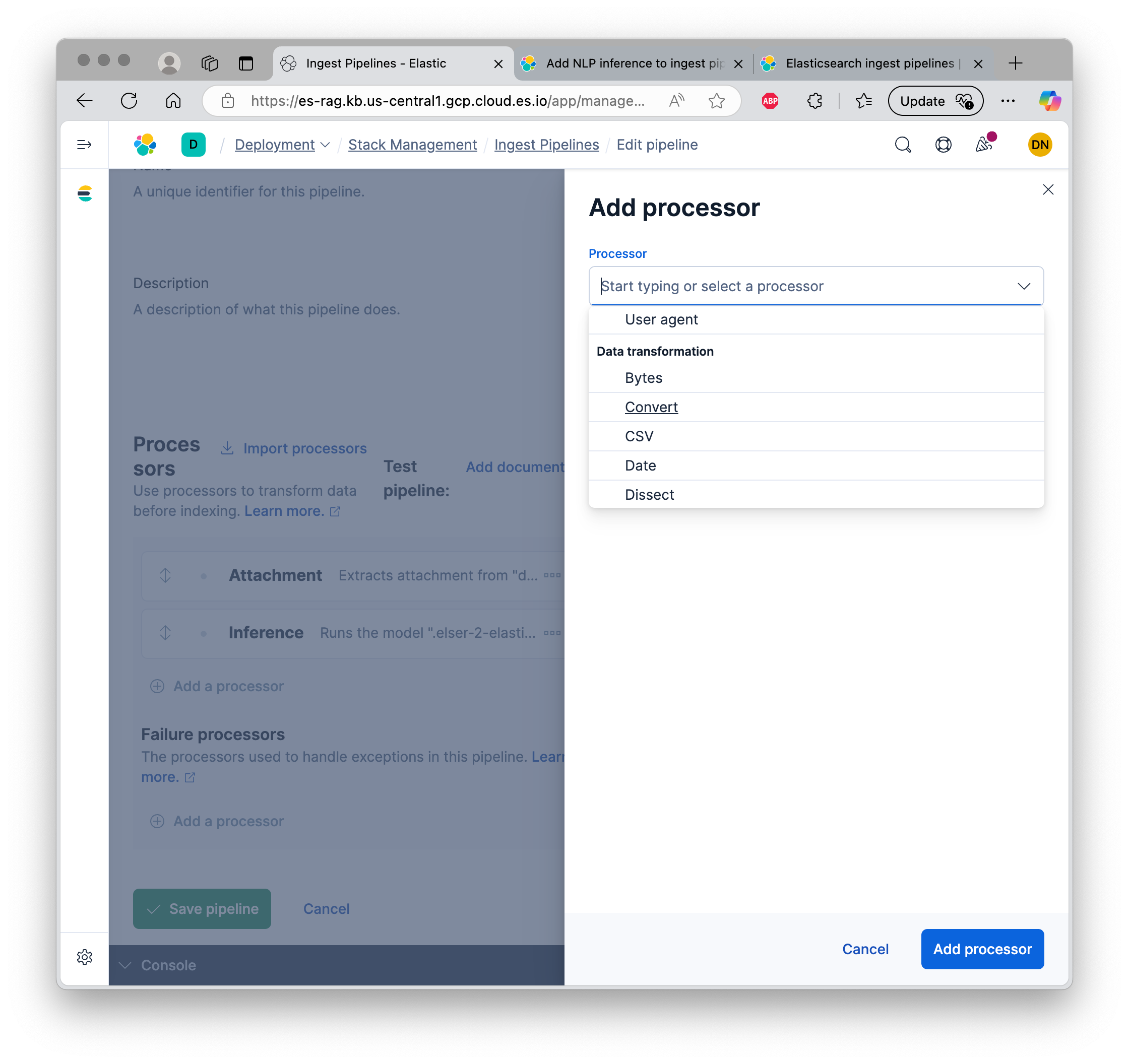
After extracting the content, we pass it through the ELSER model, asking it to create embeddings for the text in “attachment.content” field and store those embeddings in “content_embedding” field.
You can also define the pipeline graphically. The UI allows you to test the pipeline right away, either from a document within ES, or by copy-pasting JSON.
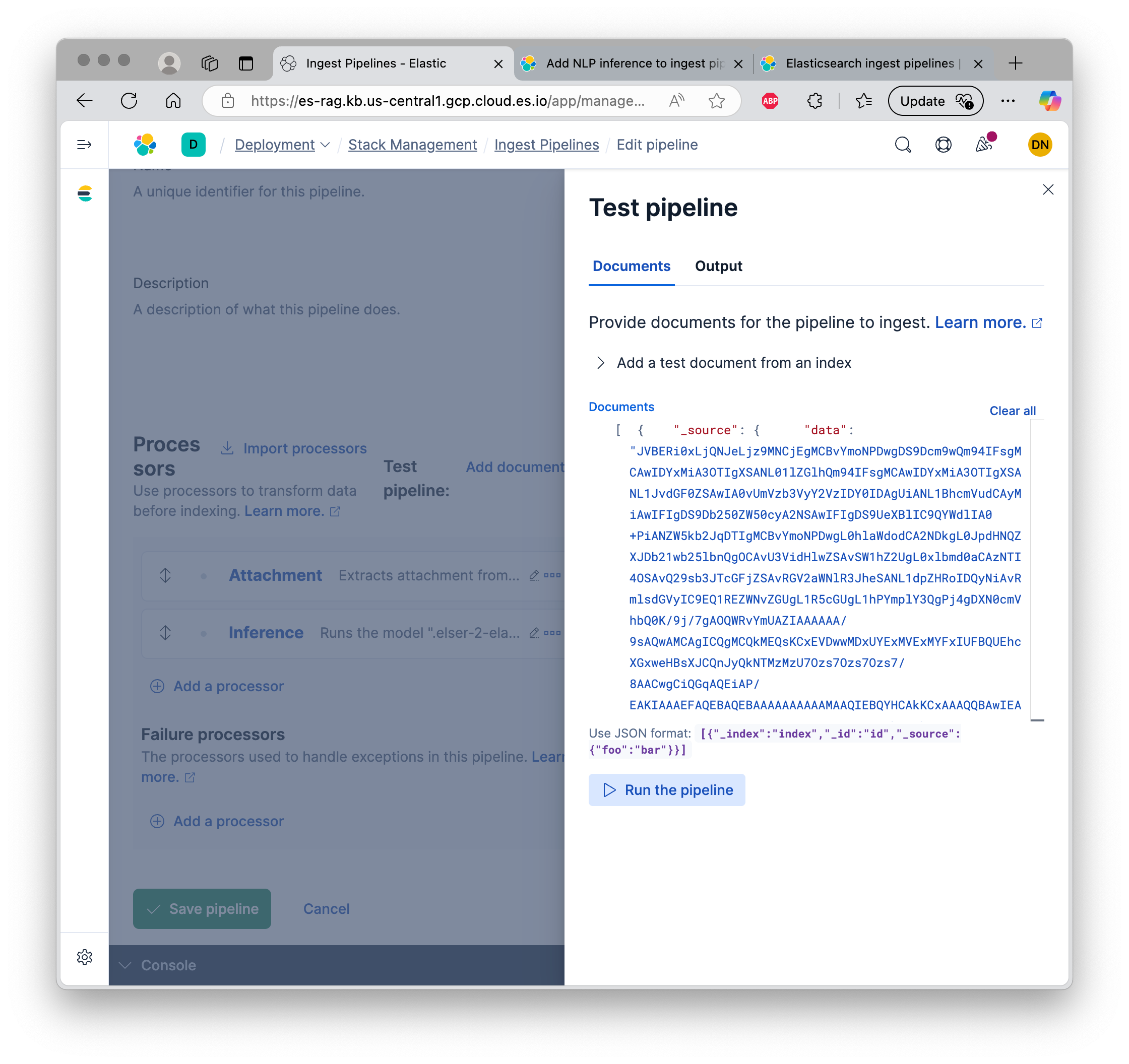
It is possible to start up ingest nodes separately to maintain ingestion throughput.
Step 2: Deploying the inference endpoint
On Elastic, under “Management”, go to “Trained Models”, then click “Deploy” on the “.elser_model_2_linux-x86_64” model. This is an auto-scaling configuration with a minimum of zero, so the first inference will be slow.
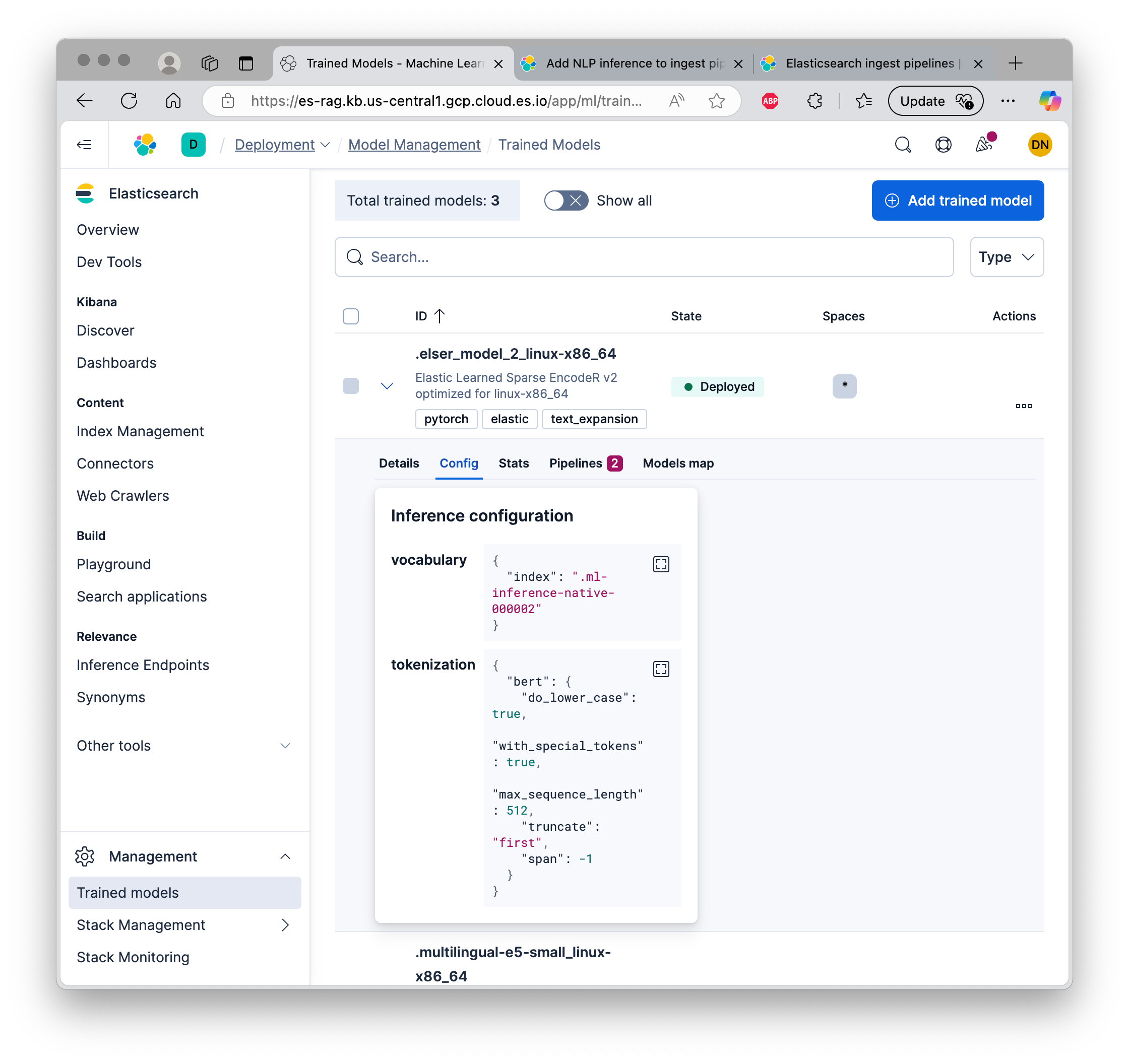
Reference documentation for ELSER.
Step 3: Defining the Index
Now that we have the ingestion pipeline defined, we can define the destination index. Elasticsearch requires us to specify the “sparse_vector” data type for where we intend to store the embeddings.
Let’s define the index.
PUT pdf-embeddings
{
"mappings": {
"properties": {
"content_embedding": {
"type": "sparse_vector"
},
"attachment.content": {
"type": "text"
}
}
}
}
Step 4: Ingesting a PDF
We can ingest a PDF from the client into our new index. This requires:
- our Elasticsearch endpoint
- an API key to run Elasticsearch ingest API
Here is a command that does that:
curl -X POST 'https://6e76ae98fe2d481bb143b75b37afbc0f.us-central1.gcp.cloud.es.io:443/pdf-embeddings/_doc?pipeline=pdf-ingest-pipeline' \
-H 'Content-Type: application/json' \
-H 'Authorization: ApiKey R3FXYVNaWUI1UzRoRkstRllpTVQ6ZVFSTHRGcVY5RXhIa0ozQVRiR1BSQQ==' \
-d '{
"data": "e1xydGYxXGFuc2kNCkxvcmVtIGlwc3VtIGRvbG9yIHNpdCBhbWV0DQpccGFyIH0="
}'
To verify that the file is ingested correctly, we can look at the index:
GET pdf-embeddings/_search
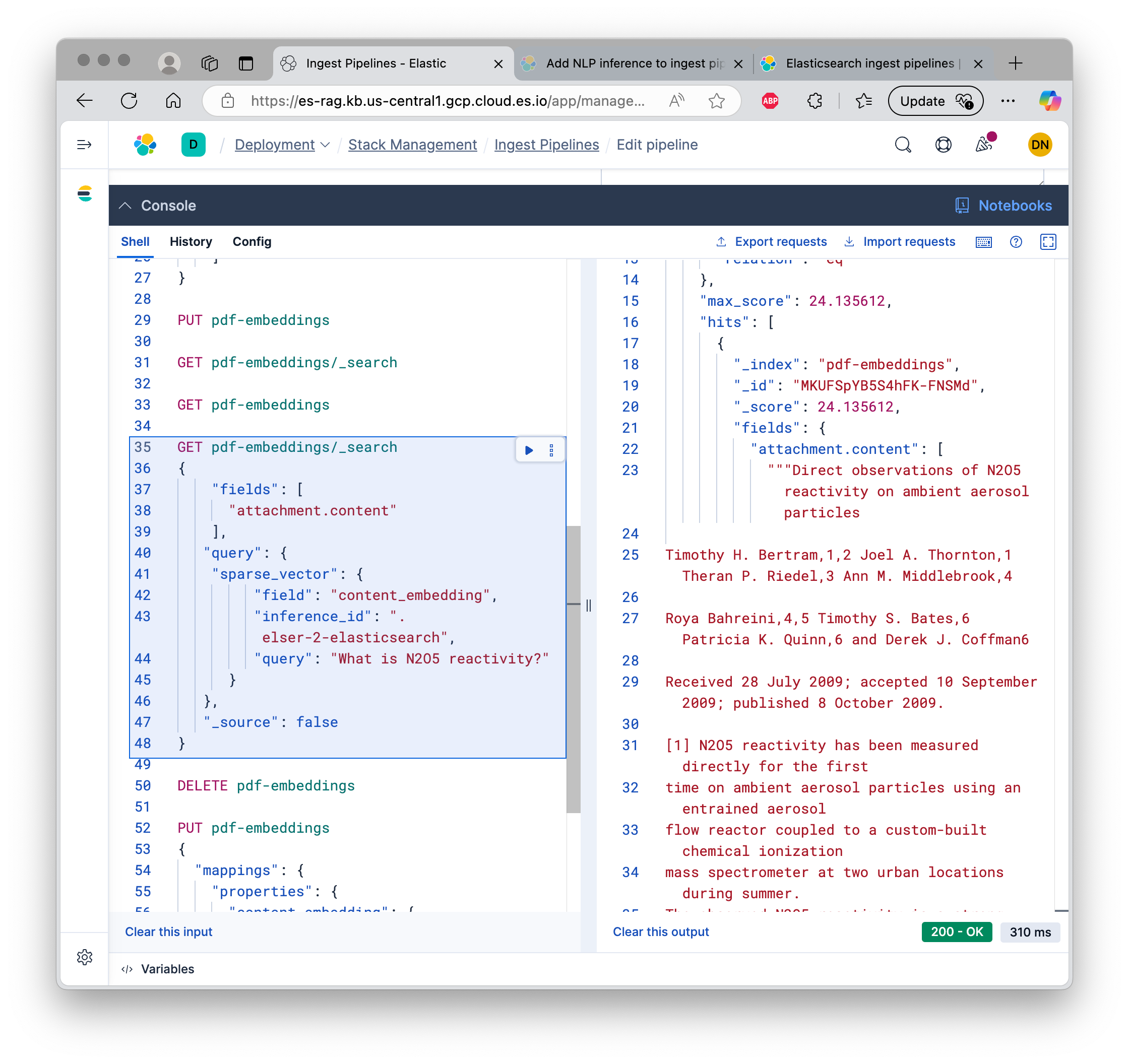
Semantically searching the PDF on ES
We can now search the PDF semantically!
GET pdf-embeddings/_search
{
"query": {
"sparse_vector": {
"field": "content_embedding",
"inference_id": ".elser-2-elasticsearch",
"query": "What is N2O5 reactivity?"
}
}
}
It uses the ELSER model again for inference, and runs a semantic search against the embeddings we have computed earlier.
Here’s the output we get. The score of 24 indicates a very good match.
{
"took": 6,
# ...
"hits": {
# ...
"max_score": 24.135612,
"hits": [
{
"_index": "pdf-embeddings",
# ...
"_score": 24.135612,
"fields": {
"attachment.content": [
"""Direct observations of N2O5 reactivity on ambient aerosol particles
Timothy H. Bertram,1,2 Joel A. Thornton,1 Theran P. Riedel,3 Ann M. Middlebrook,4
Roya Bahreini,4,5 Timothy S. Bates,6 Patricia K. Quinn,6 and Derek J. Coffman6
Received 28 July 2009; accepted 10 September 2009; published 8 October 2009.
[1] N2O5 reactivity has been measured directly for the first
# ...
"""
]
}
}
]
}
}
And the same on Elasticsearch console:
Next Steps
We can stress-test ES on a more complex pipeline that involves pre-processing on the PDF, as well as additional model calls after computing embeddings. This would simulate our precompute pipeline.
If that is successful, we can explore the cost of using Elasticsearch in this manner.